Utilizing XNA Technology in CDx Assay Development
Seeking Pharma/Biopharma/Diagnostics Partnerships
CDx tests are on the rise
28%
The overall success rate of a typical pharma drug
62%
Guided by CDx Tests
The overall success rate of a typical pharma drug
We seek partnerships with either Pharma and Biopharma or established diagnostics companies who have already partnered with Pharma or Biopharma for partnership in CDx assay development.
The use of XNA technology significantly improves the CDx assay developments
XNA technology amplifies the impact of tissue biopsy CDx, liquid biopsy CDx, and future CDx techniques such as MRD, supporting different phases of cancer care.
Affordable
The low-cost synthesis of XNA, the DNA oligo analogs, makes it more affordable than other platforms aimed at enhancing sensitivity.

Highly sensitive

Highly specific
The target-specific XNA reduces false positives by blocking the wild-type sequence amplification, as seen in the background of qPCR and ddPCR assays. XNA helps prevent patients from being chosen for ineffective treatments, saving time and money. More importantly, XNA may contribute to MRD monitoring by lowering the wild-type background.

Easy to adapt
XNA can boost sensitivity and specificity in traditional platforms like Sanger sequencing, qPCR, ddPCR, and NGS, with NO additional steps or special equipment.
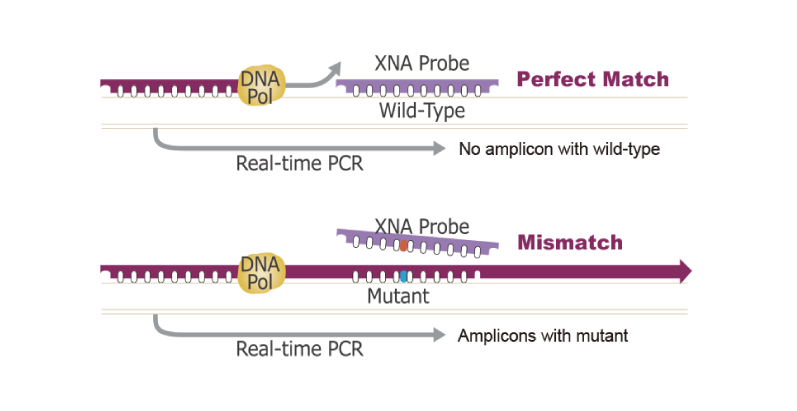
The mechanisms of the XNA technology: by blocking the wild-type sequence amplification, XNA selectively allows the amplification of mutant sequences in a PCR reaction.
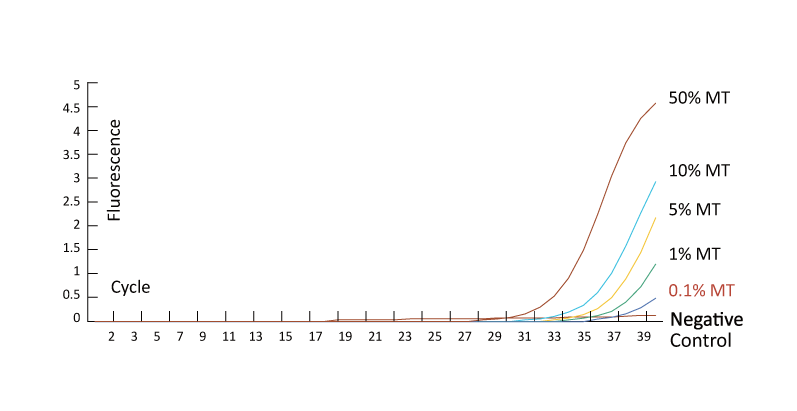
XNA blocks wild-type sequence amplification, allowing mutants with a range of Variant Allele Frequency (VAF) to be detected in a qPCR reaction, even below 0.1%.
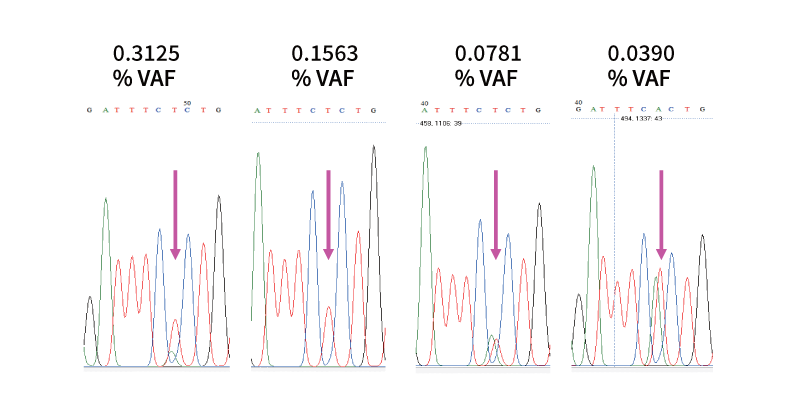
When applied in sample preparation of Sanger sequencing, XNA improves Sanger sequencing sensitivity from 15~20% to as low as 0.04%.
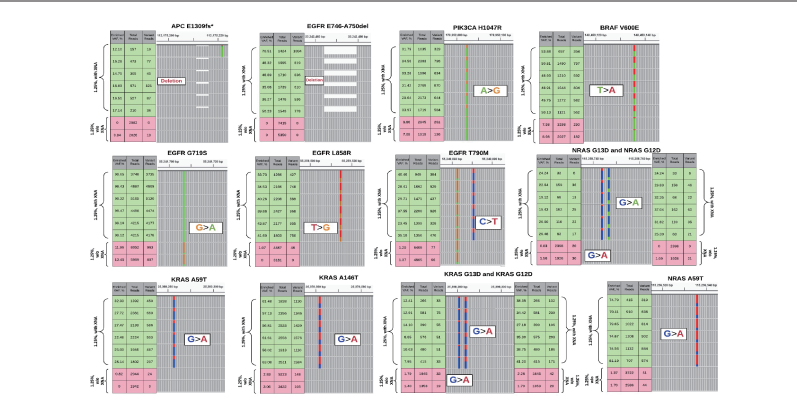
When applied to NGS, XNA significantly reduces sequencing depth for low-frequency mutation detection using plasma samples.
Affordable Excellence with Unmatched Sensitivity and Specificity
Tissue biopsy is the gold standard for clinical testing. Although many techniques are already competent for delivering sufficient sensitivity for the CDx assay using tissue biopsy, our target-specific XNA assays add value for sensitivity and specificity.
Our assays provide better accuracy by reducing potential false-negative and false-positive patient populations.
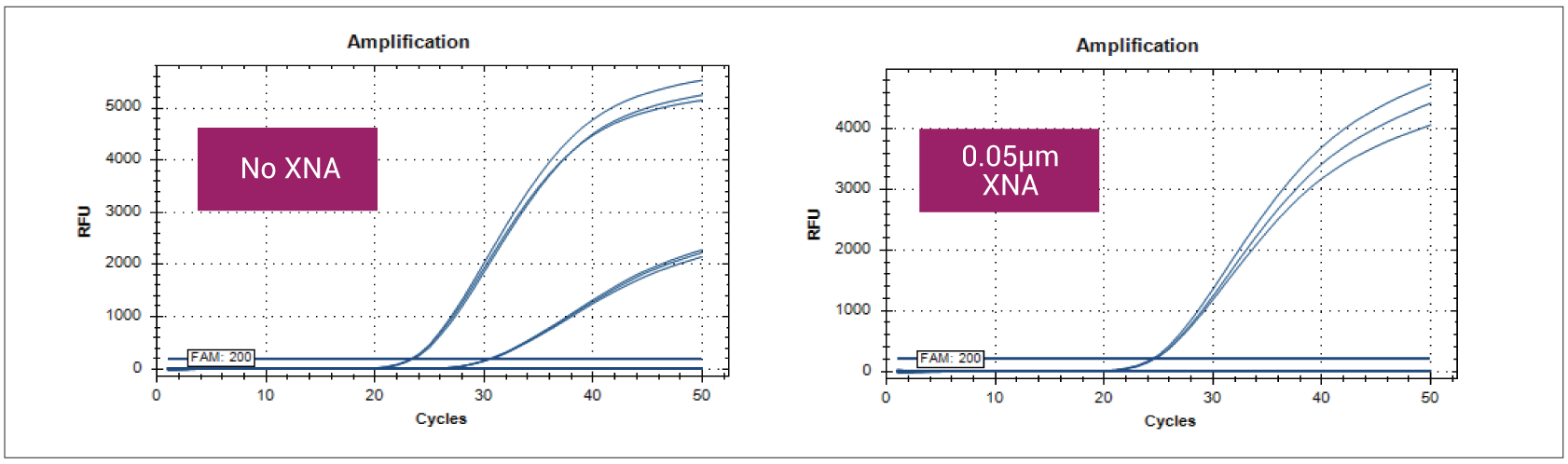
Figure: XNA is used in JAK2 V617F detection in qPCR. In a qPCR reaction, the allele-specific probe alone for JAK2 V617F fails to provide specific reactions due to the cross-reaction with the wild-type probe. However, the amplification occurs late in the cycles (In the left amplification plot, the lower curve on the right is the wild-type). However, the non-specific amplification of the wild-type is totally inhibited when a specific XNA is added to the qPCR reaction (the amplification curve on the right), increasing the assay specificity. Adding XNA has also increased the assay sensitivity from 1% to <0.05% by enriching the mutant sequence in the qPCR reaction.
Since the first FDA approval for liquid biopsy CDx in 2016, liquid biopsy has gained more traction for CDx development. The traditional biopsy is not available or difficult to get from patients for CDx-guided target therapy. Liquid biopsy can be repeatedly taken, and ctDNA (circulating tumor DNA) analyzed at different therapy stages.
With cost-effective XNA technology, the CDx tests can be used for multiple-point sample collection and detection. Adding XNA in the assay helps meet the higher-sensitivity requirement due to the low level of ctDNA available. Our XNA-based liquid biopsy assays may especially find value for cancer types with only limited ctDNA shed into the bloodstream, such as brain tumors.
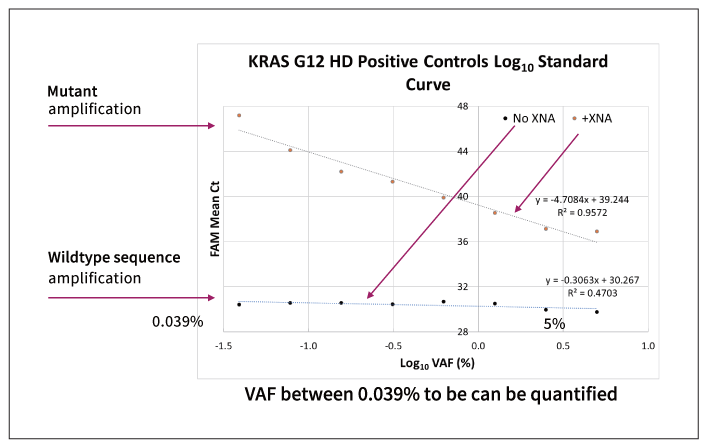
Figure 1: KRAS G12 amplification: 10 ng wildtype DNA +0.039-5% KRAS G12 mutation sequence. G12 mutation at 0.039 to 5% VAF of the mutation is detected in a qPCR reaction in the presence of target-specific XNA. Since the Ct values and the VAF are linearly correlated within this range, the VAF can be quantified. However, when no XNA is present, all the samples get amplified almost equally despite the different VAF of the KRAS G12 mutation.
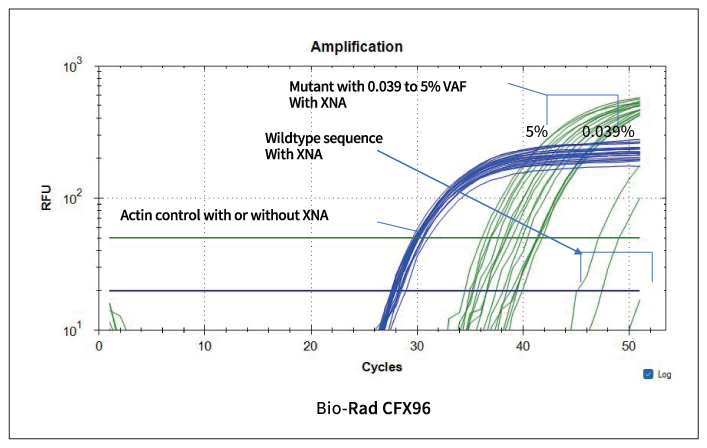
Figure 2: The amplification curve is shown. The internal control provides similar amplification, although the mutant amplification differs for the Ct value. Negative control is at high Ct value, separating from the positive control samples.
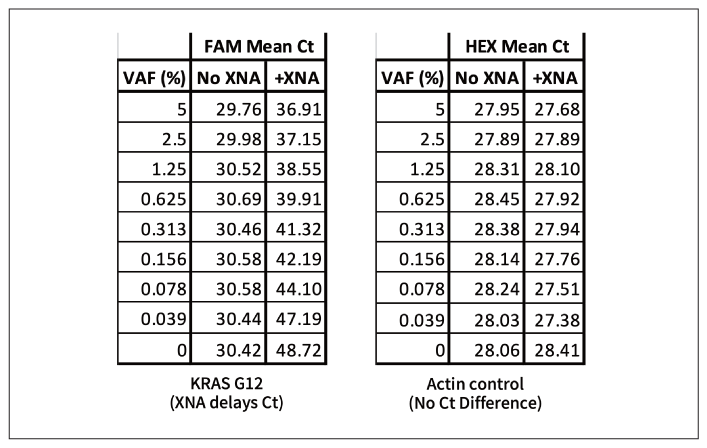
Figure 3: The Ct value is shown for both positive control and negative control in the presence or the absence of XNA. The mutant can be detected as low as 0.039% VAF.
Although MRD assays have not been approved as CDx assays, it has great potential for monitoring tumor clonal evolution. This will be important for targeted drug therapy and drug resistance therapy. Early mutation detection during the tumor clonal revolution process may provide an efficient therapy option to kill the cancer clones at the early development stage.
Target-specific XNAs can detect one copy of the variant providing a great potential for the XNA technology to be used in MRD and future CDx development.
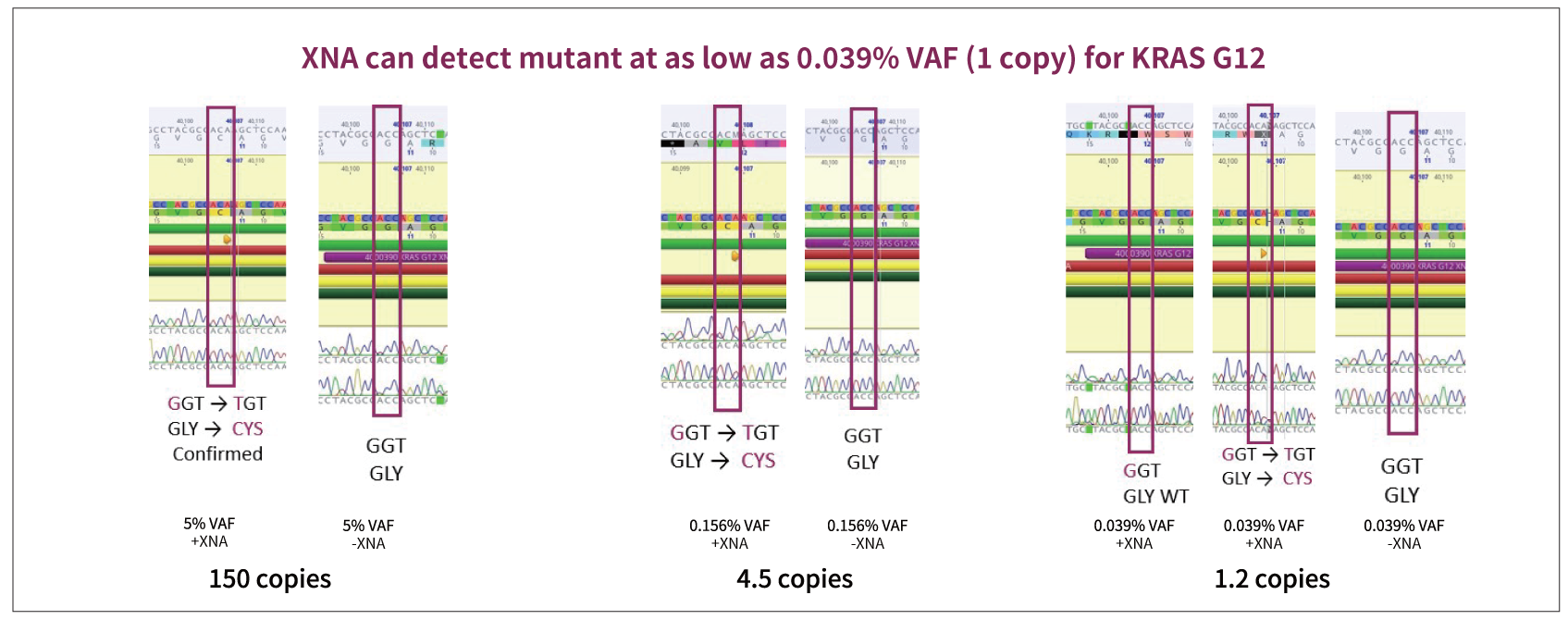
Figure: Sanger sequencing confirms KRAS G12C mutation detection from a qPCR reaction. KRAS G12 mutation (G12C) is confirmed by Sanger sequencing even at 0.039% when only 1.2 copies appear of the mutant appear in the reaction.
