Affordable Excellence with Unmatched Sensitivity and Specificity
Achieves high sensitivity and specificity by amplifying only the mutant DNA sequence while blocking amplification of the wild-type sequence, allowing to “find a needle in a haystack”.
Introducing XNA Molecular Clamp Technology
XNA, xenonucleic acids, are innovative new nucleic acid molecular oligomers that hybridize with target DNA sequences and can be employed as molecular clamps in quantitative real-time polymerase chain reactions (qPCR). The XNA tightly bind to the wild-type sequence but loosely to the mutant sequences, thereby allowing mutant sequences to be amplified in a PCR reaction while blocking wild-type sequence amplification.
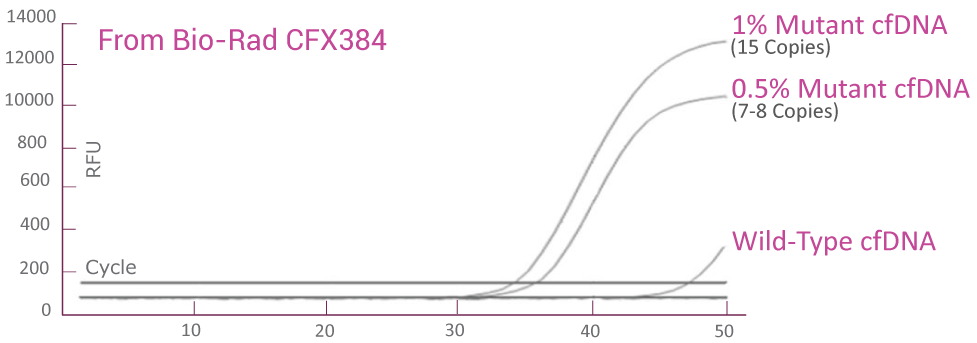
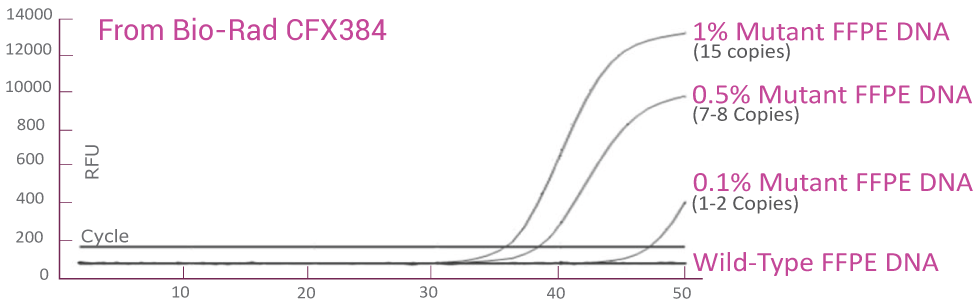
XNA molecular clamps assays can use liquid biopsy and FFPE samples to generate highly sensitive results. The limit of detection (LoD) can reach as low as 0.1% (7 or 8 copies of mutant DNA) using 5 ng of ctDNA, equivalent to 2 ml of blood from a patient.
Since the presence of circulating tumor cells (CTCs) has been found to be associated with poor survival in multiple cancers, and lowering the CTC threshold from unfavorable (≥5 CTCs/7.5 ml of blood) to favorable levels (<5 CTCs/7.5 ml of blood) improves survival, the number of CTCs in blood can be used as a predictive factor for cancer treatment response. Considering its sensitivity, the XNA technology may be used to find cancer mutations and to glean meaningful information for cancer treatment. In the context of precision medicine for cancer, finding the cancer driver mutations such as those in growth pathway and in tumor suppressor genes is critical, as it is generally recognized that these mutations contribute to the early development of different types of cancers. Detection of these mutations by sensitive XNA technologies is not only essential for early cancer detection, but for providing strategies for cancer therapy as a companion diagnostic tool. When combining with Sanger sequencing or NGS, the assay sensitivity is further increased for mutations of low variant allele frequencies (VAF).
Early Cancer detection potential by highly sensitive XNA technology
XNA molecular clamps assays are highly sensitive for use with liquid biopsy and FFPE samples.
One of the examples for cancer gene mutation detection using the XNA technology is ColoScape™ colorectal cancer mutation detection kit we developed for FFPE and liquid biopsy samples. We obtained a sensitivity of about 60% in advanced adenoma tissue samples (stage 0) compared to 42% pre-cancer detection achieved by the ColoGuard kit. For stage I to IV colon cancer, the sensitivity was 95.5%. The beauty of using XNA technology is achieving high sensitivity using regular qPCR TaqMan assays that can be performed in common pathology and genetic biomarker discovery labs without investing in other expensive instrumentation.
Analytical sensitivity in plasma (QClamp® KRAS Mutation Detection Test) powered by XNA technology
Analytical sensitivity in FFPE (QClamp® KRAS Mutation Detection Test) powered by XNA technology
Precision Medicine and Early Cancer Detection
Precision Medicine Initiative defines precision medicine as “an emerging approach for disease treatment and prevention that takes into account individual variability in genes, environment, and lifestyle for each person.” Using the precision medicine approach doctors make treatment decisions based on an individual’s genetic makeup rather than providing a generic “one-size-fits-all” solution.
In the context of precision medicine for cancer, finding the cancer driver mutations, such as those involved in growth pathway and tumor suppressor genes is critical, as it is generally recognized that these mutations contribute to early development of different types of cancers. Detection of these mutations is essential not only for early cancer detection, but also for providing strategies for using cancer therapy as a companion diagnostic tool.
The Invasive Way with FFPE Samples
Challenges for Early Cancer Detection – Invasive Method
Traditionally, tumor gene mutations have been detected using DNA isolated from formalin-fixed paraffin embedded (FFPE) tumor biopsy tissue samples. But tissue biopsy poses a few challenges:
Surgical tumor biopsies are invasive, sometimes risky, and are not always easily accessible
Mutations in a distant organ are not necessarily detected in the biopsy samples and doctors miss the opportunity to provide the appropriate treatment regimen
The dynamic mutational changes that occur with the progression of cancer are not identified by tissue biopsy, thus altering therapeutic strategies over the course of the disease becomes difficult
Challenges for Early Cancer Detection – Non-invasive Method
Recently, minimally invasive procedures, commonly called liquid biopsies, such as collecting blood samples and monitoring mutations in the circulating cell-free tumor DNA (ctDNA) in the blood, have gained traction in early cancer detection. This method has a few limitations:
Success depends on the sensitivity of the detection technologies, which becomes a limiting factor. That can also be the major reason why most of the current cancer studies and detections have been done from malignant cancers
Premaligancy studies or early cancer detection using genomic-wide analysis has been very limited due to the difficulty in detection of cancer gene mutations in a small sub-population of cells before cancer is developed
The Non-invasive Way with Liquid Biopsy
Limitations of Current Technologies for Early Cancer Detection
Finding
Source:
Genomics, Circuits, and Pathways in Clinical Neuropsychiatry, 2016
Analysis of DNA Methylation by Pyrosequencing, 2016
What is next-generation sequencing? 2013
Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data, 2017
Understanding PCR, 2017
Sanger Sequencing
Sanger sequencing is the process of selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication.
Advantage: accurate result and is, therefore, the gold standard
Disadvantage: low sensitivity (20% to 25% VAF)
NGS
Next-generation sequencing (NGS) or massively parallel or deep sequencing describes a DNA sequencing technology where millions of DNA fragments are sequenced in parallel. NGS has revolutionized genomic research making genomics easily accessible.
Advantage: high-throughput and good sensitivity (1% to 5% VAF, or even better)
Disadvantage: costly and time-consuming (7 to 10 days)
qPCR Analysis
Quantitative polymerase chain reaction (qPCR) is a primer-probe-based method by which the amount of the PCR product can be determined in real-time and is very useful for investigating gene expression
Advantage: Sensitivity can reach 1% VAF for some targets. Rapid with minimal hands-on work.
Disadvantage: Multiple methods available for qPCR and a lot of variations in sensitivity. Some of them are only 10% VAF
Pyrosequencing Assays
Pyrosequencing is a technique that uses a sequencing-by-synthesis system, designed to quantify single-nucleotide polymorphisms (SNPs).
Advantage: better sensitivity and throughput than Sanger sequencing; the early form of NGS assays
Disadvantage: low sensitivity (5% to 8% VAF)
ddPCR
Droplet Digital PCR (ddPCR) technology is a method of performing PCR in a sample that is fractionated into thousands of water-oil emulsion droplets. It utilizes Taq polymerase in a standard PCR reaction to amplify a target DNA fragment in each individual droplet using pre-validated primer or primer/probe assays.
Advantage: high sensitivity (claimed to be 0.001% VAF)
Disadvantage: much less sensitivity observed in testing than claimed and suffers from false-positive results
XNA Technology
Sensitivity reaches 0.1% to 0.5% VAF; much more sensitive than regular qPCR and other qPCR-derived techniques.
Achieves ultra-sensitivity by amplifying only mutant DNA and blocking wild-type sequences.






