isobDNA™ isoC-isoG Branched DNA Technology
High-throughput and sensitive mutation detection and quantification capability without DNA sequencing
Qualitative and Quantitative DNA Copy or
Gene Expression Analysis
isobDNA™ technology uses isoC-isoG Branched DNA Technology to qualitatively or quantitatively measure the presence of target DNA or RNA. It has been used in FDA-approved clinical applications, including prognosis and monitoring of patients with viral diseases. Unlike PCR or RT-PCR, isobDNA™ technology does not amplify the DNA or RNA in the samples but amplifies the signals for detection. Because the signal amplified is proportional to the levels of the target (DNA or RNA) in the samples, the latter is easy to be quantified with a standard curve.
Advantages of isobDNA™ Technology Compared with RT-PCR
Although widely used and approved as powerful technologies, PCR and RT-PCR have a few drawbacks when used as molecular diagnostics tools.
PCR and RT-PCR rely on amplification of DNA or cDNA to generate signals for diagnostic analysis and amplified DNA often has cross-contamination problems without proper operational control. isobDNA™ technology does not have this problem
A direct comparison of RT-PCR and branched DNA technology has shown them to correlate well in determining target gene expression of HIV. However, RT-PCR showed greater proportional system errors and was shown to be less efficient for quantitation of some subtype of HIV virus. At low target concentration, RT-PCR was better but with increase in target concentration, branched DNA technology showed more reliable results.
View the comparative study on branched DNA and RT-PCR assays
The Principle and Workflow for isobDNA™ Technology
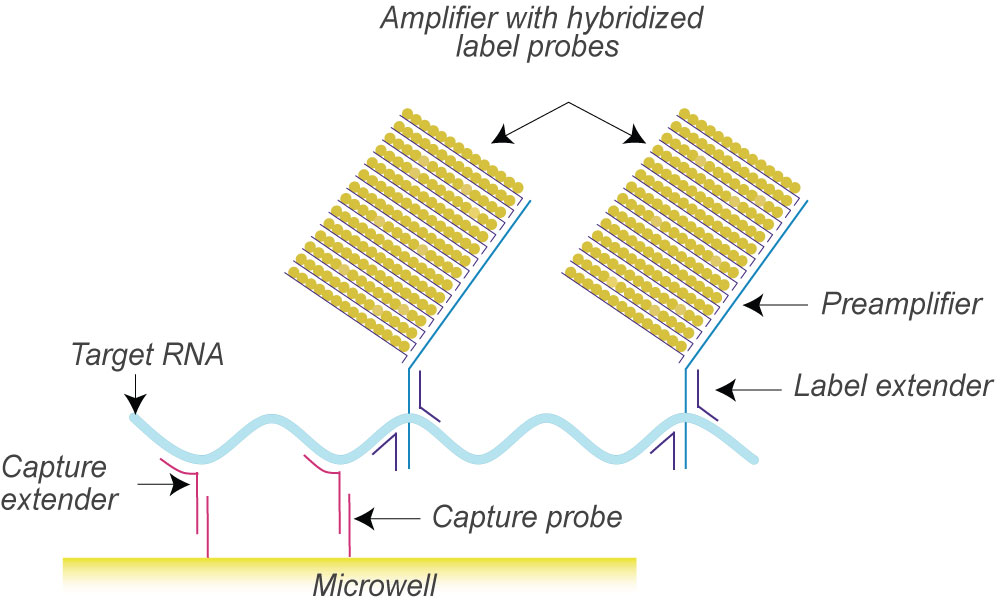
isobDNA™ technology uses a series of hybridization events to capture the target and another series of hybridization events to connect the labeled probes for chemiluminescent signal amplification. Such a “sandwich” structure efficiently captures and quantifies the target in the samples with a signal amplification of 500 to 10 million molecules.
First, “capture probes” that are pre-coated on the bottom of plates or conjugated to beads or discs hybridize with “target probes” that bind to specific target DNA or RNA sequences. Once the samples are added, the target DNA or RNA sequence is captured and the rest of the samples are washed away. Next, a second set of probes, including “preamplifier” and “amplifier” probes labeled with alkaline phosphatases are added to hybridize with the target molecules to form the “sandwich”. The second wash washes away the unhybridized probes. Finally, the substrate for alkaline phosphatase is added and the ensuing chemiluminescent signals are detected. The workflow for the detection is illustrated blow.
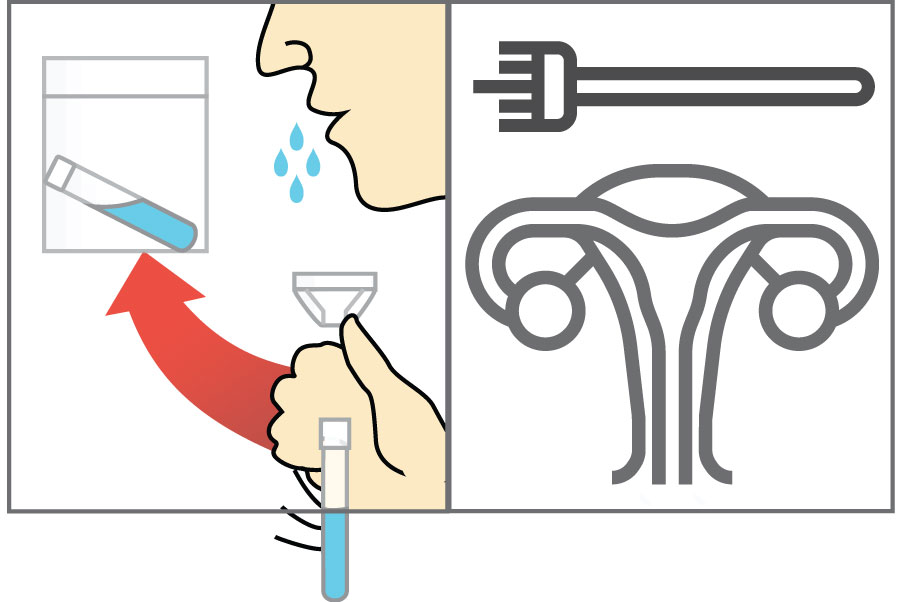
Step 1: Lyse Samples
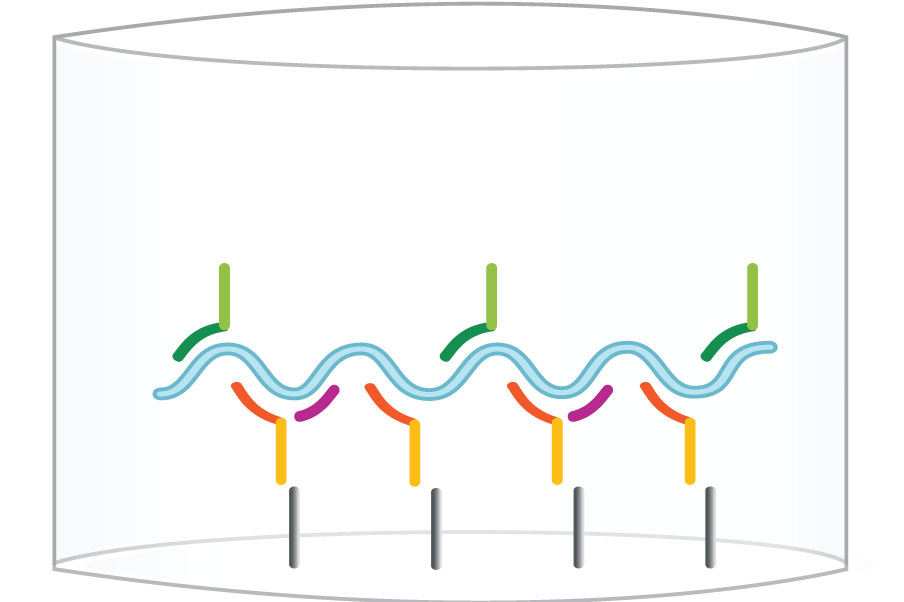
Step 2: Capture E6/E7
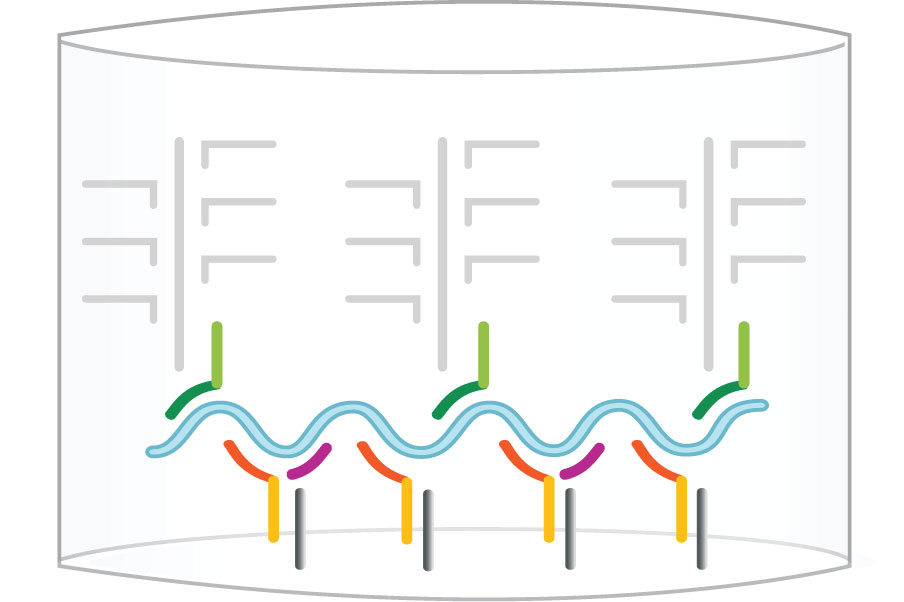
Step 3: Amplifier
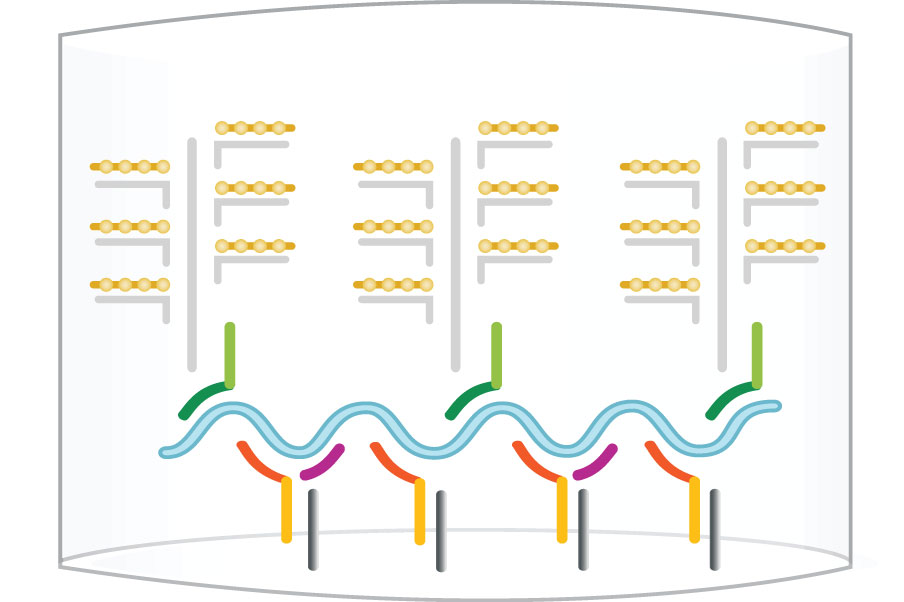
Step 4: Label Probe - AP
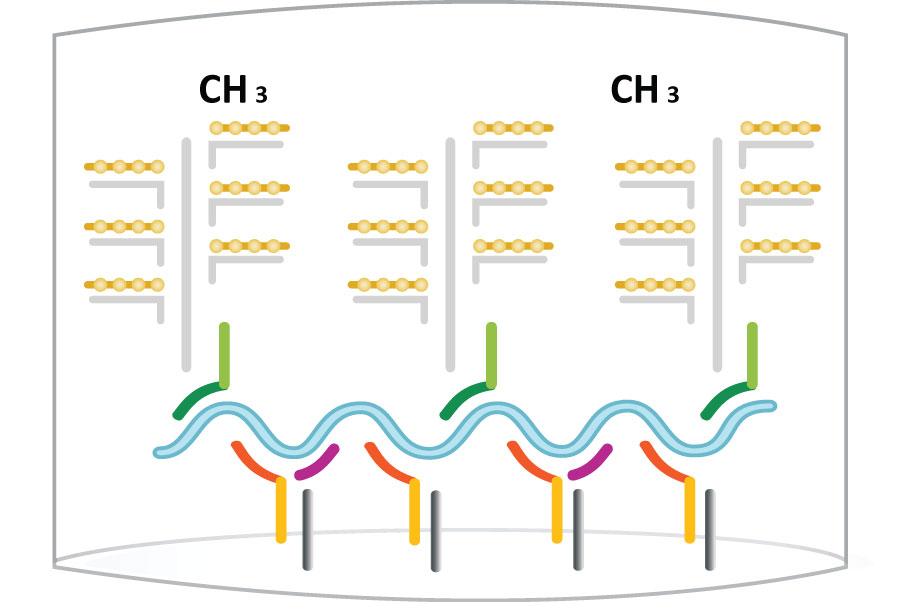
Step 5: Substrates

Step 6: Read and Report
isobDNA™ Technology Platform: Single Assay or Multiplex Assay?
isobDNA™ technology can be used both in a single well assay format for a specific target or as multiplex assays for multiple targets in the discs or beads format. Contact us for the different formats available for your assay.
Applications for isobDNA™ Technology
isobDNA™ technology can be used in different areas for target DNA or RNA detection, including pathogen detection, biomarker quantitation, gene expression, and in situ hybridization. The benefits of these assays can be summarized as shown below:
Gene Expression Analysis
Ideal clinical platform for companion diagnostic delivery
Low cost and rapid turn-around time
96-well format. No RNA purification required
Highly sensitive: detects 50 transcripts per ml
Pathogen Detection (Virus, Fungi and Bacteria)
No requirement for target purification and amplification
Improved sensitivity and specificity
In Situ Hybridization
Single molecule detection
FFPE and frozen tissue compatible
Strong/bright signal with low background
Multiple-4-plex easily attainable
An ideal complement to antibody-based advance tissue staining compatible with bright field and fluorescent microscopy
isobDNA™ Featured Application: QuantiVirus™ HPV E6/E7 mRNA Test
QuantiVirus™ HPV E6/E7 mRNA Test – Predicting Pre-Cancer in Cervical Cancer and Head & Neck Cancer Screening
Human papillomavirus (HPV), with over 40 distinct HPV types, is the most common sexually transmitted infection in the U.S., affecting 42.5% of the adults aged 18–59 years during 2013–2014. The risk for persistent infection with some HPV types is the development of cervical cancer for women and head and neck cancer in men.
QuantiVirus™ HPV E6/E7 mRNA test detects HPV oncogene E6/E7 mRNA from 14 high-risk types. It
isobDNA™ Featured Application: Radtox™ cfDNA Test
RadTox™ cfDNA Test – Radiation Therapy Toxicity Monitoring
Powered by isobDNA™ Technology, RadTox™ cfDNA Test is developed for cancer patients’ radiation therapy toxicity monitoring. The test fulfills an unmet medical need in cancer radiation therapy by enabling the direct monitoring of the severity of side effects and tumor response within days of radiation treatment initiation. RadTox™ would allow dose escalation to tumors in less-sensitive patients, aid in the development of radiotoxicity mitigators, and reduce overtreatment risk for sensitive patients. The assay provides a fast, cost-effective quantitation of liquid biopsy cfDNA sample from cancer patients before and after radiation therapy and provides patient information for estimates of dosage use and general radiation resistance.
