QClamp® NRAS Mutation Detection Tests
Improved Sensitivity for Single Gene Mutation Detection
The QClamp® NRAS Mutation Detection Test for plasma and Formalin-Fixed Paraffin-Embedded (FFPE) samples aids in the identification of patients eligible for cancer treatment and in monitoring response to therapy, which can lead to improved outcomes in cancer patients. The QClamp® NRAS Mutation Detection Test is an in vitro diagnostic real-time quantitative PCR assay for the detection of somatic mutations in and near codons 12 and 13 in Exon 2, codons 59 and 61 in Exon 3, and codons 117 and 146 in Exon 4 in the human NRAS gene, using purified DNA extracted from FFPE or plasma.
Powered by XNA technology, the QClamp® NRAS Mutation Detection Test has achieved a much higher analytical sensitivity compared to other commercial qPCR kits and other cancer gene mutation detection methods. QClamp® NRAS Mutation Detection Test is able to detect reliably 0.1% to 0.5% mutant DNA out of wild-type DNA for targeted mutations, providing lower detection limit compared to similar assays available in the market due to robust enrichment of mutant sequences while suppressing amplification of wild-type sequences.
Product Catalog
QClamp® NRAS Mutation Detection Test – CE Version: DC-10-3020
QClamp® NRAS Mutation Detection Test – Research-Use Version: DC-10-3020R
Pack Size: 30 Samples
Customized Kits Offering
Pick one or more existing NRAS target from the standard NRAS kit: Codons 12, 13, 59, 61, 117 and 146
Service Offering
We provide research service for QClamp® NRAS Mutation Detection Test.
GET A QUOTE NOW
Advantages of QClamp® NRAS Mutation Detection Test

ULTRA-SENSITIVE
Reliably detects 0.1% to 0.5% VAF mutant DNA out of wild-type DNA for targeted mutations

SAMPLE READY

LOW INPUT DNA
Minimum 5ng input DNA per reaction. Less than 2 tubes of blood (10mL each) needed for cfDNA

COMPREHENSIVE COVERAGE
Covering all relevant somatic mutations in 6 codons of NRAS oncogene
FAST RESULTS
Less than 4 hours of assay run time

GREAT VERSATILITY
Validated on the most common qPCR machines with minimized variability
NRAS Mutation and Cancer
Introduction of NRAS Gene
NRAS, like KRAS and HRAS, is a member of the RAS family of GTPases and plays a central role in the MAPK signaling pathway. NRAS activating mutations in codons 12, 13, and 61 have been found in various cancers, including melanoma (15-20%), colorectal cancer (1-6%), lung cancer (1%), hepatocellular carcinoma (10%), myeloid leukemia (14%), and thyroid carcinoma (7%). Mutations in the NRAS gene generate resistance to anti-NRAS therapeutic agents such as Cetuximab and to chemotherapy, and are prognostic indicators of poor survival in patients with metastatic colorectal cancer (mCRC).
NRAS Gene Detection May Benefit the Immunotherapy
With increased understanding of the utilization of the immune system to combat cancer, immune therapy has now become the mainstay for treatment of advanced melanoma. It has been suggested that patients with NRAS mutations benefit better from immunotherapies compared to other mutations. Detection of NRAS mutations may provide useful information for immunotherapy strategies.
Supporting Data for QClamp® NRAS Mutation Detection Test
QClamp® NRAS Mutation Detection Test analytical sensitivity in plasma: detects as low as 0.125% VAF mutant DNA in a 5ng input
cfDNA extracted from
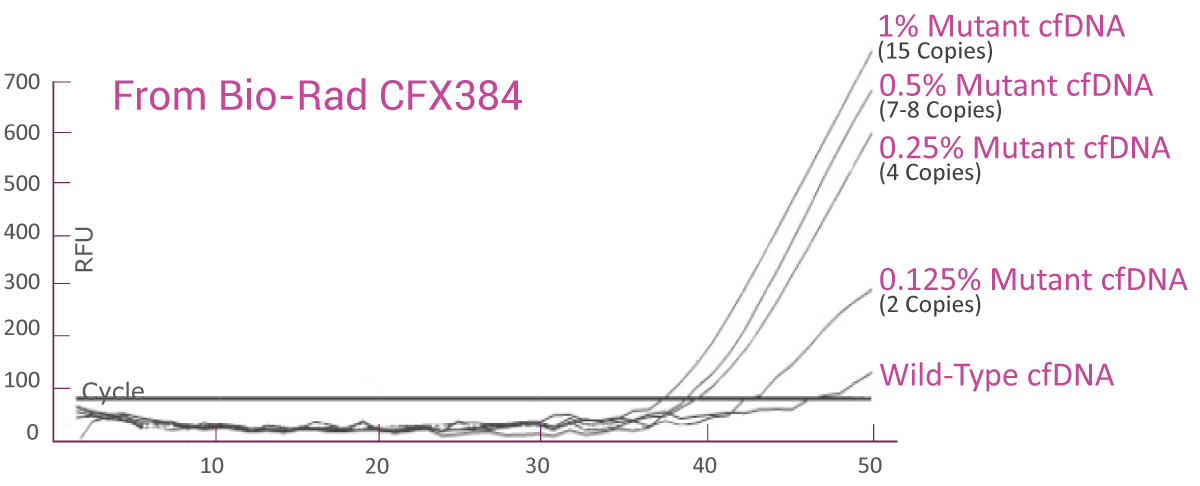
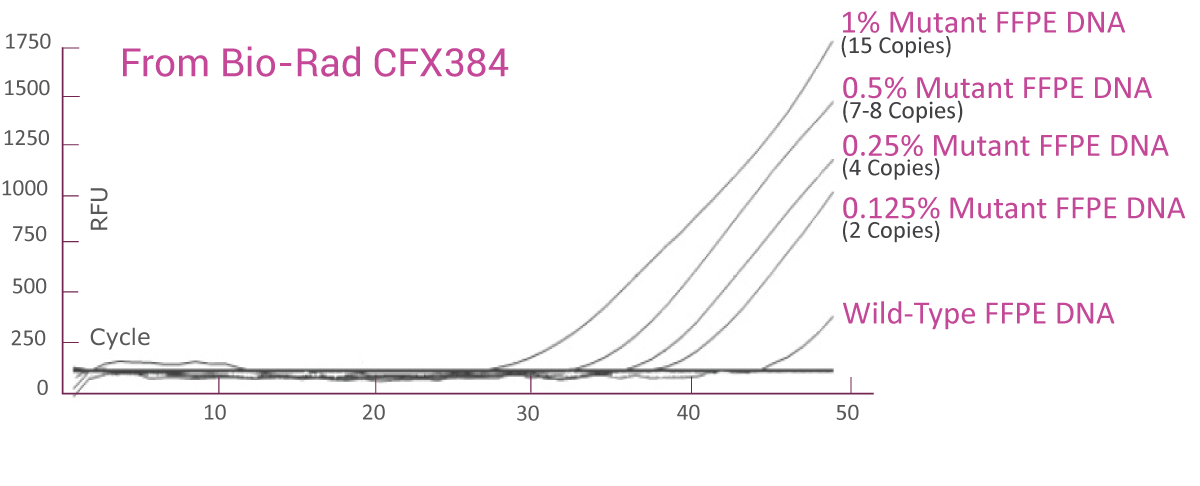
QClamp® NRAS Mutation Detection Test analytical sensitivity in FFPE: detects as low as 0.125% VAF mutant DNA in a 5ng input
DNA extracted from FFPE of a colorectal cancer patient with known NRAS G12D mutation was diluted in 5ng/μl of wild-type FFPE DNA. VAF of the sample was verified by NGS. The NRAS c12 assay was able to detect as low as 2 NRAS G12D mutant gene copies (0.125% VAF) in the background of wild-type FFPE DNA.
Streamlined Workflow for QClamp® Gene Mutation Detection Tests

Step 1: DNA Isolation & Quantification
Extract DNA from FFPE or plasma using a commercial DNA extraction kit and measure the concentration using fluorometric analysis

Step 2: set up qpcr
Mix the assay reagents, load into PCR plate, add controls and extracted DNA ~ 30-60 minutes
Step 3: Amplification parameters
Enter amplification parameters on
qPCR instrument, load PCR plate
and start the run ~ 2.5 hours
Step 4: Data analysis
Determine the presence or absence
of mutations according to the Cq
value cutoffs ~ 15 minutes
Resources
Catalog Number
Pack Size: 10-sample
CE catalog #: DC-10-2020
Research-use-only (RUO) catalog #: DC-10-3020R
Pack Size: 30-sample
CE catalog #: DC-10-3020
Research-use-only (RUO) catalog #: DC-10-3020R
Pack Size: 60-sample
CE catalog #: DC-10-4020
Research-use-only (RUO) catalog #: DC-10-4020R
Detected Codons
Codons 12, 13, 59, 61, 117 and 146
Sample Type
Plasma and FFPE
Input DNA
5-10ng/Reaction
Validated Instruments
Roche LightCycler® 480, Bio-Rad CFX384 and ABI QuantStudio 5
Detection Channel
FAM; HEX
Detection Chemistry
TaqMan
Turnaround Time
Less than 4 hours
Stability
Stable for 12 months at -25 ℃ to -15 ℃
Most frequent NRAS mutations detected by QClamp® NRAS Mutation Detection Test
| Exon | Amino Acid Change | Nucleotide change | Cosmic No. |
|---|---|---|---|
| 2 | p.G12D | c.35G>A | 564 |
| p.G12S | c.34G>A | 563 | |
| p.G12C | c.34G>T | 562 | |
| p.G12V | c.35G>T | 566 | |
| p.G12A | c.35G>C | 565 | |
| p.G12R | c.34G>C | 561 | |
| p.G12N | c.34_35GG>AA | 12723 | |
| p.G12G | c.36T>C | 567 | |
| p.G12P | c.34_35GG>CC | 559 | |
| p.G12Y | c.34_35GG>TA | 560 | |
| p.G12E | c.35_36GT>AG | 144577 | |
| p.G13D | c.38G>A | 573 | |
| p.G13R | c.37G>C | 569 | |
| p.G13V | c.38G>T | 574 | |
| p.G13C | c.37G>T | 570 | |
| p.G13A | c.38G>C | 575 | |
| p.G13S | c.37G>A | 571 | |
| p.G13G | c.39T>C | 576 | |
| p.G13N | c.37_38GG>AA | 24668 | |
| p.G13Y | c.37_38GG>TA | 568 | |
| p.G13V | c.38_39GT>TC | 572 | |
| 3 | p.A59T | c.175G>A | 578 |
| p.A59D | c.176C>A | 253327 | |
| p.A59fs*4 | c.174_175ins13 | 5022497 | |
| p.A59S | c.175G>T | 5351687 | |
| p.A59G | c.176C>G | 5878737 | |
| p.Q61R | c.182A>G | 584 | |
| p.Q61K | c.181C>A | 580 | |
| p.Q61L | c.182A>T | 583 | |
| p.Q61H | c.183A>T | 585 | |
| p.Q61H | c.183A>C | 586 | |
| p.Q61P | c.182A>C | 582 | |
| p.Q61E | c.181C>G | 581 | |
| p.Q61R | c.181_182CA>AG | 579 | |
| p.Q61Q | c.183A>G | 587 | |
| p.Q61L | c.181_182CA>TT | 12725 | |
| p.Q61K | c.180_181AC>TA | 12730 | |
| p.Q61L | c.182_183AA>TG | 30646 | |
| p.Q61R | c.182_183AA>GG | 33693 | |
| p.Q61* | c.181C>T | 6005482 | |
| p.Q61T | c.181_182CA>AC | 5044300 | |
| p.Q61K | c.181_183CAA>AAG | 53223 | |
| p.Q61_E62>HK | c.183_184AG>CA | 26494 | |
| 4 | p.K117N | c.351A>T | |
| p.A146T | c.436G>A | 27174 | |
| p.A146V | c.437C>T | 4170228 | |
Download
For products that are in stock, DiaCarta will arrange shipment in 1-3 days. For products that are on backorder, DiaCarta will arrange shipment in 3-5 weeks.
Intended Use: QClamp® NRAS Mutation Detection Test is CE marked. Outside the
Shipping Condition: QClamp® NRAS Mutation Detection Test will be shipped with dry ice. For domestic shipment, DiaCarta provides overnight delivery through FedEx Domestic Overnight Shipping Service. For international shipment, DiaCarta provides 3-7 days in transit through FedEx International Priority Shipping Service. Please contact DiaCarta if you prefer to use your shipping carrier.
